热门标签
热门文章
- 1@Autowired注入失败_@autowired 不成功
- 2图(Graph)详解 - 数据结构_图数据结构
- 3Xilinx FPGA原语总结_obufds原语
- 4Spring中Bean的作用域与生命周期_been的生命周期
- 5C++ LibCurl 库的使用方法_libcurl c++
- 6基于Java的连连看游戏设计与实现_java连连看
- 7ORA-12514: TNS: 监听程序当前无法识别连接描述符中请求服务的解决方法_ora12514监听程序当前无法识别连接描述符中请求的服务
- 8Android Studio——记事本案例_android studio记事本
- 9清华学霸直博简历火了!CPU、操作系统、编译器全自主写,刘知远点赞
- 10Flutter,更高效、更实用的跨端开发选择_fluter开发跨平台
当前位置: article > 正文
Medical SAM Adapter: Adapting SegmentAnything Model for Medical Image Segmentation_medical sam adapter: adapting segment anything mod
作者:笔触狂放9 | 2024-06-30 18:32:50
赞
踩
medical sam adapter: adapting segment anything model for medical image segme
Junde Wu
1
,
2
,
3
,
5
, Rao Fu
1
, Yu Zhang
1
, Huihui Fang
2
,
3
, Yuanpei Liu
4
, Zhaowei
Wang
5
, Yanwu Xu
2
,
3
In this paper, instead of finetuning the SAM model, we propose Med SAM Adapter, which integrates the medical specific domain knowledge to the segmentation model, by a simple yet effective adaptation technique.
在本文中,我们提出了Med SAM适配器,而不是微调MAM适配器,它通过一种简单而有效的自适应技术,将医学特定领域的知识集成到分割模型中。
Introduction
简介
The main reason for SAM’s failure over medical images is due to the lack of training data. Although SAM established a sophisticated and efficient data engine in the training, they collected few cases for medical usage. In this paper, we attempt to expand SAM towards prompt-based medical image segmentation with minimum effort. Technically, we choose to fine-tune the pre-trained SAM using a parameter-efficient fine-tuning (PEFT) technique called Adaption.
SAM在医疗图像上失败的主要原因是缺乏训练数据。尽管SAM在培训中建立了一个复杂和高效的数据引擎,但他们收集的医疗使用案例很少。在本文中,我们试图将SAM扩展到以最小努力的基于提示的医学图像分割。从技术上讲,我们选择使用一种称为自适应的参数高效微调(PEFT)技术来对预先训练过的SAM进行微调。
接下来介绍了为什么选择这个方法的理由,可以详见论文。
Method
方法
Preliminary: SAM architecture 这一部分先对SAM结构进行了回顾,详见论文以及相关文献
Instead of fully turning all parameters, we kept the pre-trained SAM parameters frozen and inserted an Adapter module at specific positions in the architecture.
我们没有完全翻转所有参数,而是将预先训练过的SAM参数冻结,并在体系结构中的特定位置插入一个适配器模块。
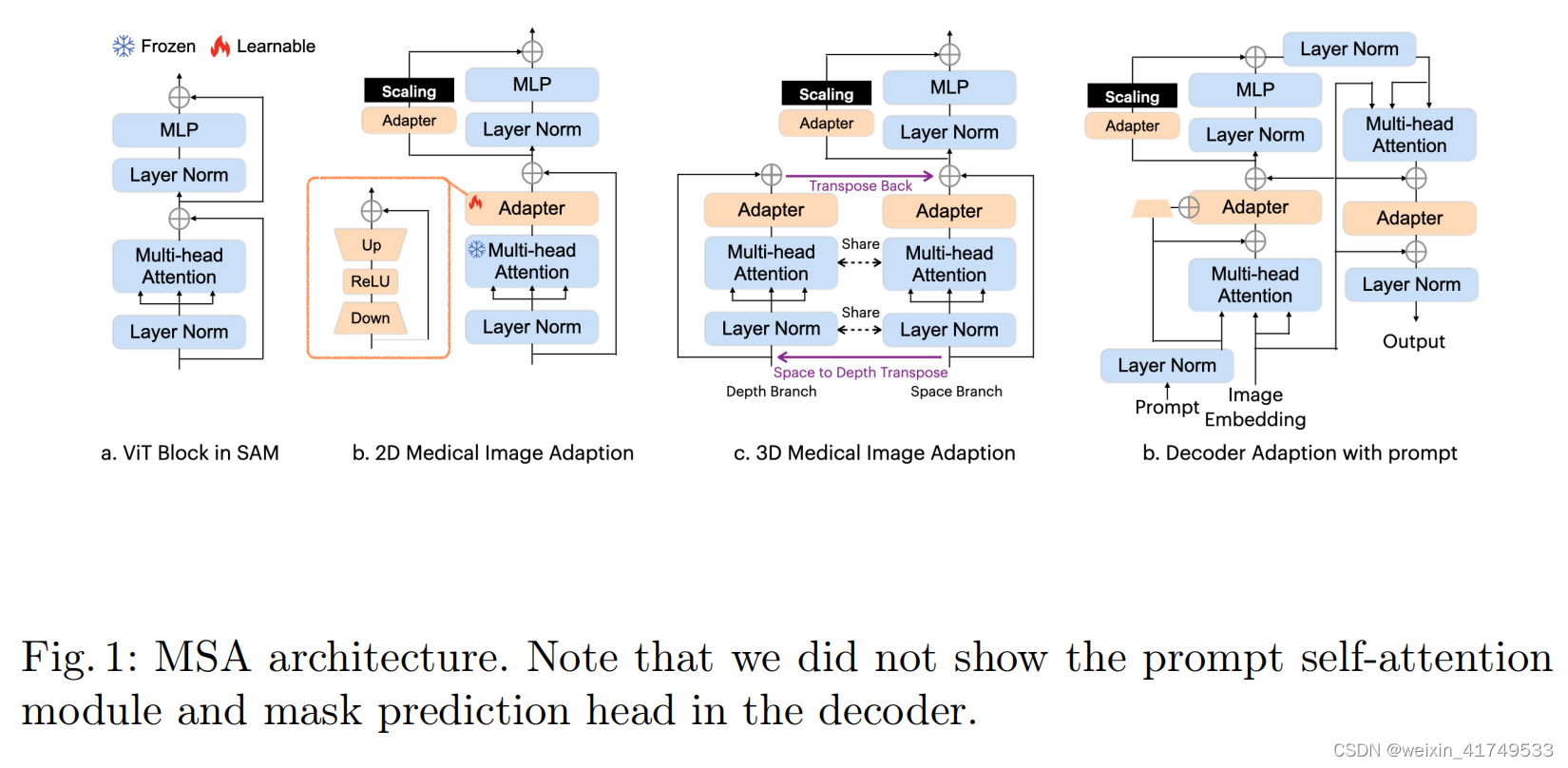
In the SAM encoder, we have deployed two adapters for each ViT block. For a standard ViT block (as shown in Fig. 2 (a)), we have placed the first Adapter after the multi-head attention and before the residue connection (as shown in Fig. 2 (b)), and the second Adapter in the residue path of the MLP layer after the multi-head attention. Immediately after the second Adapter, we have scaled
the embedding with a scale factor
s
following
在SAM编码器中,我们已经为每个ViT块部署了两个适配器。对于一个标准的ViT块(如图2(a)所示),我们将第一个适配器放置在多头注意之后和残留连接之前(如图2(b)所示),第二个适配器放置在多头注意后的MLP层的剩余路径上。在第二个适配器之后,我们立即用比例因子s对嵌入进行了缩放
In the SAM decoder, we have deployed three adapters for each ViT block. The first Adapter is deployed after the prompt-to-image embedding multi-head cross attention with a residue addition of the prompt embedding. We have made a small modification on the Adapter to integrate the prompt information into the module. Specifically, we have used another down projection to compress the prompt embedding, and added it into the Adapter over the embedding before ReLU activation. This modification helps the Adapter to turn the parameter conditioned on the prompt information and be more flexible and general to different modalities and downstream tasks. The second Adapter in the decoder is deployed in exactly the same way as in the encoder, to adapt the MLP enhanced embedding. The third Adapter is deployed after the residue connection of the image embedding-to-prompt cross-attention. Another residue connection and layer normalization are connected after the adaption to output the final results. Note that we have only deployed the Adapter in the first block of the two blocks of the Decoder. The second block and the mask prediction head are fully turned over the given data.
在SAM解码器中,我们已经为每个ViT块部署了三个适配器。第一个适配器是在提示到图像嵌入多头交叉注意后部署的,并添加提示嵌入。我们对适配器做了一个小的修改,以将提示信息集成到模块中。具体来说,我们使用了另一个向下投影来压缩提示嵌入,并在ReLU激活之前将其添加到嵌入之上的适配器中。这种修改有助于适配器以提示信息为条件,对不同的模式和下游任务更灵活和通用。解码器中的第二个适配器的部署方式与编码器中的部署方式完全相同,以适应mlp增强的嵌入。第三个适配器是在图像嵌入到提示交叉注意的剩余连接后部署的。自适应后连接另一个剩余连接和层规范化,输出最终结果。请注意,我们只在解码器的两个块的第一个块中部署了适配器。第二个块和掩模预测头被完全翻转给定的数据。
we propose a novel adaptation method inspired by the image-to-video adaptation, with some modifications. The specific architecture is shown in Fig. 2 (c). In each block, we split the attention operation into two branches: the space branch and the depth branch. For a given 3D sample with depth D, we send D x N x L to the multi-head attention in the space branch, where N is the number of embeddings, and L is the length of the embedding. Here, D is the number of operations, and the interaction is applied over N x L to learn and abstract space correlations as embeddings. In the depth branch, we first transpose the input matrix to obtain N x D x L and then send it to the same multi-head attention. Although we use the same attention mechanism, the interaction is applied over D x L. In this way, depth correlations are learned and abstracted. Finally, we transpose the results from the depth branch back to their original shape and add them to the results of the space branch
为解决医疗图像中三维图像连续层相关性的问题,我们提出了一种新的适应方法,启发图像到视频的适应,并进行了一些修改。具体架构如图2 (c).所示在每个区块中,我们将注意力操作分为两个分支:空间分支和深度分支。对于给定的深度为D的三维样本,我们将D x N x L发送给空间分支中的多头注意,其中N为嵌入数,L为嵌入的长度。这里,D是操作的次数,交互被应用于N x L来学习和抽象空间相关性作为嵌入。在深度分支中,我们首先将输入矩阵转置得到N x D x L,然后将其发送给同一多头注意。虽然我们使用相同的注意机制,但交互作用被应用在D x L上。通过这种方式,深度相关性被学习和抽象。最后,我们将来自深度分支的结果转置回它们原来的形状,并将它们添加到空间分支的结果中。
Training Strategy
训练策略
预训练:
Instead of the MAE pretraining used in SAM, we use a combination of several self-supervised learning methods for pre-training. The first two are Contrastive Embedding-Mixup (e-Mix) and Shuffled Embedding Prediction (ShED), following [32]. e-Mix is a contrastive objective that additively mixes a batch of original input embeddings, weighting them with different coefficients. It then trains an encoder to produce a vector for a mixed embedding that is close to the original inputs’ embeddings in proportion to their mixing coefficients. ShED shuffles a fraction of embeddings and trains the encoder with a classifier to predict which embeddings were perturbed. We also use a Masked Autoencoder (MAE) following the original implementation of SAM, which masks a given fraction of input embeddings and trains models to reconstruct them.
训练用数据来源于医学影像公开数据集,详见论文,对模型进行预训练。我们没有在SAM中使用MAE预训练,而是结合使用几种自监督学习方法进行预训练。前两个方法是对比嵌入混合(e-Mix)和打乱嵌入预测(ShED),遵循[32]。e-Mix是一个对比目标,它同时混合一批原始输入嵌入,用不同的系数进行加权。然后,它训练一个编码器为混合嵌入产生一个向量,该向量接近于原始输入的嵌入,与它们的混合系数成比例。ShED对部分嵌入进行洗牌,并使用分类器训练编码器来预测哪些嵌入受到了扰动。我们还根据SAM的原始实现使用了一个屏蔽的自动编码器(MAE),它屏蔽了给定的输入嵌入部分,并训练模型来重建它们。
Training with Prompt
使用prompt训练
We use a different text prompt training strategy with SAM. In SAM, the authors used the image embedding of the target object crop produced by CLIP as the image embedding close to its corresponding text description or definition in CLIP. However, since CLIP is barely trained on medical image datasets, it can hardly relate the organs/lesions on the image with the corresponding text definition. Instead, we first randomly generate several free texts containing the
definition of the target (i.e., optic disc, brain tumor) as the keyword from ChatGPT, and then extract the embedding of the text using CLIP as the prompt for training. One free text could contain multiple targets, in which case we supervise the model with all their corresponding masks
我们首先使用随机抽样进行初始化,然后使用迭代抽样过程添加一些点击。我们对SAM使用了不同的文本提示训练策略。在SAM中,作者将CLIP产生的目标对象作物的图像嵌入作为接近CLIP中对应的文本描述或定义的图像嵌入。然而,由于CLIP几乎没有在医学图像数据集上进行训练,因此它很难将图像上的器官/病变与相应的文本定义联系起来。相反,我们首先从ChatGPT中随机生成几个包含目标定义(即视盘、脑瘤)作为关键字的免费文本,然后使用CLIP作为训练提示,提取文本的嵌入。一个自由文本可以包含多个目标,在这种情况下,我们用它们所有对应的面具来监督模型。
Result
实验结果
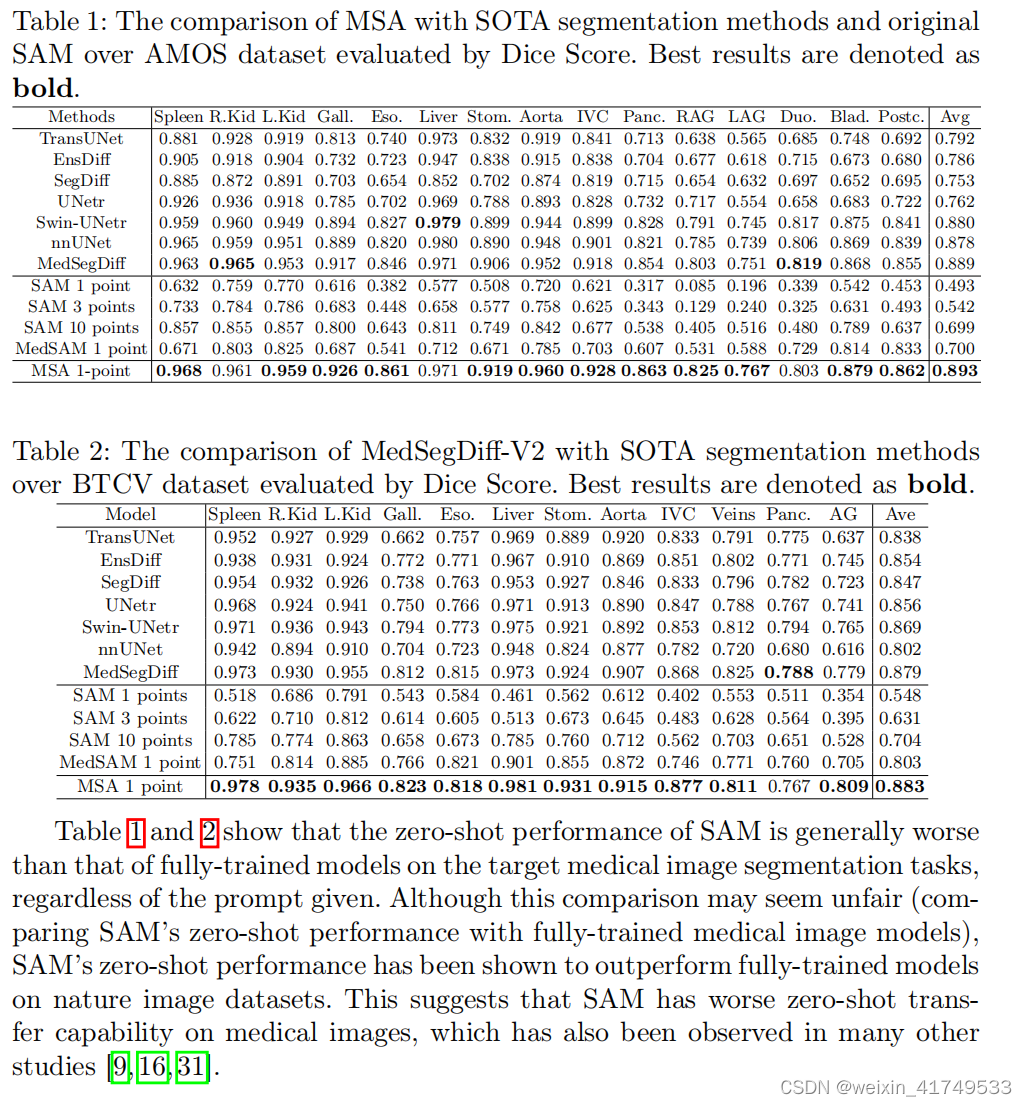
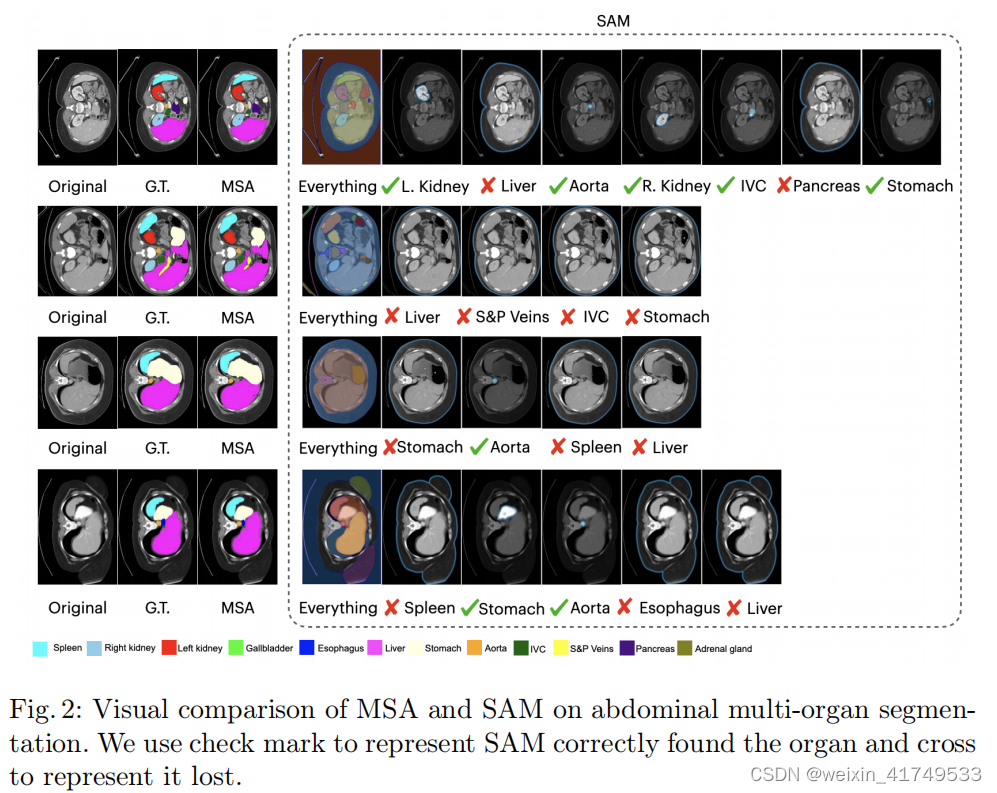
声明:本文内容由网友自发贡献,不代表【wpsshop博客】立场,版权归原作者所有,本站不承担相应法律责任。如您发现有侵权的内容,请联系我们。转载请注明出处:https://www.wpsshop.cn/w/笔触狂放9/article/detail/773587
推荐阅读
相关标签


