热门标签
当前位置: article > 正文
遗传算法C语言实现以及思路详解简单易懂_c语言实现遗传算法
作者:IT小白 | 2024-07-08 09:17:38
赞
踩
c语言实现遗传算法
前言
前几天刚用python实现了遗传算法用于求解函数最值,详见遗传算法详解python代码实现以及实例分析然后我又用c语言写了一遍,因为当时老师说只能用c语言,但是他发的实验报告里又说能用python、matlab了,挺奇怪的,没事,写都写完了,分享一下吧。
首先讲一下用c语言和python写这题的一些区别,由于c语言相对于python来说比较底层,所以涉及到的复杂的操作会比较少,但是由于c比python快不止一倍两倍,基本就不用咋考虑性能了,怎么好理解我就怎么写了。
python用一个numpy矩阵就可以表示一个种群了,c语言我是用一个animal结构体数组来表示种群;
python在进行优胜劣汰的时候,是根据适应度转换为对应被抽取的概率然后用numpy.random.choice根据概率进行对应抽取形成新的种群,而c语言里我是通过计算出适应度概率之后计算ceil(适应度概率 * 种群数量),然后以此作为此animal被留下的数量。
主要是一些细节的不同,整体思路是完全一致的。
实例代码解释
1.初始化参数以及变量设置
#include<bits/stdc++.h> using namespace std; # define Int_bit 3 // 整数占的bit位 # define DNA_bit 16 // 一个DNA的二进制位数,(第一维表示符号位) # define DNA_num 2 // DNA的个数 # define animal_num 200 // 开始种群的数量 # define cross_rate 0.8 // 生殖交叉概率 # define variation_rate 0.005 // 变异的概率 # define generator_n 100 // 种群演变的次数 int limit_area[2] = {0, 10}; // 值域 struct animals{//存储x, y两个二进制表示 int DNA[DNA_bit * DNA_num]; }animal[animal_num], selected_animal[animal_num]; struct DNA_results{//二进制转化为十进制的结果存储 double translated_result[DNA_num]; }DNA_result[animal_num]; struct fitnesses{//每个animal的适应度计算 double p; int id; }fitness[animal_num];
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
参数设置和python那篇是一样的。
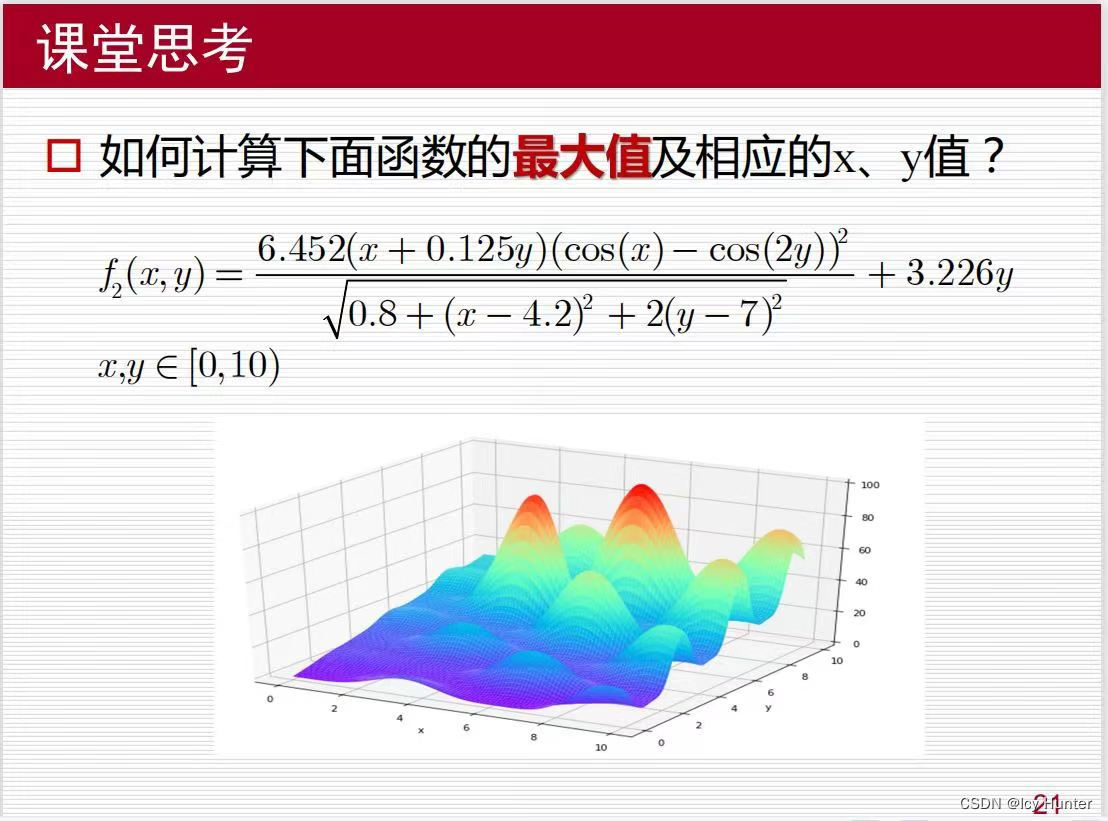
2.定义环境(定义目标函数)
double f(double x, double y){
return (6.452 * (x+0.125*y) * pow(cos(x)-cos(2*y), 2)) / (pow(0.8+pow(x-4.2, 2) + 2*pow(y-7, 2), 0.5))+ 3.226*y;
}
- 1
- 2
- 3
3.DNA解码(计算x,y)
void DNA2to10(int n){//将二进制解码为十进制 for(int i=0; i<DNA_num; i++){ double sum = 0; int base = i * DNA_bit; double sign = animal[n].DNA[base]; double flag; if(sign == 0)flag = -1; else flag = 1; for(int j=base+1; j<=base+Int_bit; j++){ if(animal[n].DNA[j] == 1)sum += pow(2, Int_bit+base-j); } for(int j=base+Int_bit+1; j<DNA_bit+base; j++){ if(animal[n].DNA[j] == 1)sum += pow(2, Int_bit+base-j); } DNA_result[n].translated_result[i] = sum * flag; } }
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
4.初始化种群(初始化解,考虑定义域)
srand((unsigned)time(NULL));//时间随机
for(int i=0; i<animal_num; i++){//初始化种群
for(int j=0; j<DNA_bit * DNA_num; j++){
animal[i].DNA[j] = rand()%2;
}
}
for(int i=0; i<animal_num; i++){//如果有不符合定义域的就再生成一遍
for(int j=0; j<DNA_bit * DNA_num; j++){
animal[i].DNA[j] = rand()%2;
}
if(flag_limit_area(limit_area, i) != 1)i-=1;
}
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
5.计算适应度(计算误差,考虑定义域)
//适应度排序的方法(带着id然后按适应度排序 int cmp(fitnesses f1, fitnesses f2){ return f1.p < f2.p; } void get_fitness(){//计算每个animal的适应度 double fitness_score[animal_num]; double fit_flag[animal_num]; for(int i=0; i<animal_num; i++){ DNA2to10(i); fitness_score[i] = f(DNA_result[i].translated_result[0], DNA_result[i].translated_result[1]); fit_flag[i] = flag_limit_area(limit_area, i); } double minn=8888888; for(int i=0; i<animal_num; i++)minn = min(minn, fitness_score[i]); for(int i=0; i<animal_num; i++){ fitness_score[i] = fitness_score[i] - minn + 1e-5; } double sum = 0; for(int i=0; i<animal_num; i++){ fitness_score[i] = fitness_score[i] * fit_flag[i]; sum += fitness_score[i]; }; for(int i=0; i<animal_num; i++){ fitness[i].p = fitness_score[i] / sum; fitness[i].id = i; } }
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
6.适者生存(挑选误差较小的答案)
void select(){//根据适应度选择animal
int n = 0;
for(int i=animal_num; i>=0; i--){
int num = ceil(fitness[i].p * animal_num);
for(int j=0; j<num; j++){
for(int k=0; k<DNA_bit * DNA_num; k++){
selected_animal[n].DNA[k] = animal[fitness[i].id].DNA[k];
}
n++;
if(n == animal_num)break;
}
if(n == animal_num)break;
}
}
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
7.生殖、变异(更改部分二进制位,取反部分二进制位,可能生成误差更小的答案)
void crossover_and_variation(){//生殖交叉、变异 for(int k=0; k<animal_num; k++){ int father[DNA_bit * DNA_num]; int mother[DNA_bit * DNA_num]; int child[DNA_bit * DNA_num]; int father_id = rand() % 200; int mother_id = rand() % 200; for(int i=0; i<DNA_bit * DNA_num; i++){ father[i] = animal[father_id].DNA[i]; mother[i] = animal[mother_id].DNA[i]; } if((rand()%100)/100.0 < cross_rate){ int cross_pos = rand() % (DNA_bit * DNA_num); for(int i=0; i<cross_pos; i++)child[i] = father[i]; for(int i=cross_pos; i<DNA_bit * DNA_num; i++)child[i] = mother[i]; } if((rand()%100)/100.0 < variation_rate){ int variation_pos = rand() % (DNA_bit * DNA_num); child[variation_pos] = 1 - child[variation_pos]; } for(int j=0; j<DNA_bit * DNA_num; j++){ selected_animal[k].DNA[j] = child[j]; } } }
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
8.copy函数(将选择的selected_animal赋值回animal以便迭代遗传进化)
void copy(){//将新的结果copy回animal以便进行下一轮迭代
for(int i=0; i<animal_num; i++){
for(int j=0; j<DNA_bit * DNA_num; j++){
animal[i].DNA[j] = selected_animal[i].DNA[j];
}
}
}
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
9.遗传进化以及结果选择
for(int i=0; i<generator_n; i++){//遗传进化 get_fitness();//适应度计算 sort(fitness, fitness+animal_num, cmp);//排序,方便挑选 select();//挑选种群 copy();//将挑选好的复制回原来的种群 crossover_and_variation();//交配、变异 copy();//将生成的子种群复制回原来的种群 } //得出最终的最优结果 get_fitness(); sort(fitness, fitness+animal_num, cmp); int id = fitness[animal_num-1].id; double x = DNA_result[id].translated_result[0]; double y = DNA_result[id].translated_result[1]; cout<<"最优结果:"<<endl; cout<<"x: "<<x<<" "<<"y: "<<y<<endl; cout<<f(x, y);
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
10.完整代码
#include<bits/stdc++.h> using namespace std; # define Int_bit 3 // 整数占的bit位 # define DNA_bit 16 // 一个DNA的二进制位数,(第一维表示符号位) # define DNA_num 2 // DNA的个数 # define animal_num 200 // 开始种群的数量 # define cross_rate 0.8 // 生殖交叉概率 # define variation_rate 0.005 // 变异的概率 # define generator_n 100 // 种群演变的次数 int limit_area[2] = {0, 10}; // 值域 struct animals{//存储x, y两个二进制表示 int DNA[DNA_bit * DNA_num]; }animal[animal_num], selected_animal[animal_num]; struct DNA_results{//二进制转化为十进制的结果存储 double translated_result[DNA_num]; }DNA_result[animal_num]; struct fitnesses{//每个animal的适应度计算 double p; int id; }fitness[animal_num]; int cmp(fitnesses f1, fitnesses f2){ return f1.p < f2.p; } double f(double x, double y){ return (6.452 * (x+0.125*y) * pow(cos(x)-cos(2*y), 2)) / (pow(0.8+pow(x-4.2, 2) + 2*pow(y-7, 2), 0.5))+ 3.226*y; } void DNA2to10(int n){//将二进制解码为十进制 for(int i=0; i<DNA_num; i++){ double sum = 0; int base = i * DNA_bit; double sign = animal[n].DNA[base]; double flag; if(sign == 0)flag = -1; else flag = 1; for(int j=base+1; j<=base+Int_bit; j++){ if(animal[n].DNA[j] == 1)sum += pow(2, Int_bit+base-j); } for(int j=base+Int_bit+1; j<DNA_bit+base; j++){ if(animal[n].DNA[j] == 1)sum += pow(2, Int_bit+base-j); } DNA_result[n].translated_result[i] = sum * flag; } } int flag_limit_area(int limit_area[], int i){//检测是否超过定义域 DNA2to10(i); if(DNA_result[i].translated_result[0] >= limit_area[0] && DNA_result[i].translated_result[0] <= limit_area[1] && DNA_result[i].translated_result[1] >= limit_area[0] && DNA_result[i].translated_result[1] <= limit_area[1]) return 1; else return 0; } void get_fitness(){//计算每个animal的适应度 double fitness_score[animal_num]; double fit_flag[animal_num]; for(int i=0; i<animal_num; i++){ DNA2to10(i); fitness_score[i] = f(DNA_result[i].translated_result[0], DNA_result[i].translated_result[1]); fit_flag[i] = flag_limit_area(limit_area, i); } double minn=8888888; for(int i=0; i<animal_num; i++)minn = min(minn, fitness_score[i]); for(int i=0; i<animal_num; i++){ fitness_score[i] = fitness_score[i] - minn + 1e-5; } double sum = 0; for(int i=0; i<animal_num; i++){ fitness_score[i] = fitness_score[i] * fit_flag[i]; sum += fitness_score[i]; }; for(int i=0; i<animal_num; i++){ fitness[i].p = fitness_score[i] / sum; fitness[i].id = i; } } void select(){//根据适应度选择animal int n = 0; for(int i=animal_num; i>=0; i--){ int num = ceil(fitness[i].p * animal_num); for(int j=0; j<num; j++){ for(int k=0; k<DNA_bit * DNA_num; k++){ selected_animal[n].DNA[k] = animal[fitness[i].id].DNA[k]; } n++; if(n == animal_num)break; } if(n == animal_num)break; } } void crossover_and_variation(){//生殖交叉、变异 for(int k=0; k<animal_num; k++){ int father[DNA_bit * DNA_num]; int mother[DNA_bit * DNA_num]; int child[DNA_bit * DNA_num]; int father_id = rand() % 200; int mother_id = rand() % 200; for(int i=0; i<DNA_bit * DNA_num; i++){ father[i] = animal[father_id].DNA[i]; mother[i] = animal[mother_id].DNA[i]; } if((rand()%100)/100.0 < cross_rate){ int cross_pos = rand() % (DNA_bit * DNA_num); for(int i=0; i<cross_pos; i++)child[i] = father[i]; for(int i=cross_pos; i<DNA_bit * DNA_num; i++)child[i] = mother[i]; } if((rand()%100)/100.0 < variation_rate){ int variation_pos = rand() % (DNA_bit * DNA_num); child[variation_pos] = 1 - child[variation_pos]; } for(int j=0; j<DNA_bit * DNA_num; j++){ selected_animal[k].DNA[j] = child[j]; } } } void copy(){//将新的结果copy回animal以便进行下一轮迭代 for(int i=0; i<animal_num; i++){ for(int j=0; j<DNA_bit * DNA_num; j++){ animal[i].DNA[j] = selected_animal[i].DNA[j]; } } } int main(){ srand((unsigned)time(NULL));//时间随机 for(int i=0; i<animal_num; i++){//初始化种群 for(int j=0; j<DNA_bit * DNA_num; j++){ animal[i].DNA[j] = rand()%2; } } for(int i=0; i<animal_num; i++){//如果有不符合定义域的就再生成一遍 for(int j=0; j<DNA_bit * DNA_num; j++){ animal[i].DNA[j] = rand()%2; } if(flag_limit_area(limit_area, i) != 1)i-=1; } for(int i=0; i<generator_n; i++){//遗传进化 get_fitness();//适应度计算 sort(fitness, fitness+animal_num, cmp);//排序,方便挑选 select();//挑选种群 copy();//将挑选好的复制回原来的种群 crossover_and_variation();//交配、变异 copy();//将生成的子种群复制回原来的种群 } //得出最终的最优结果 get_fitness(); sort(fitness, fitness+animal_num, cmp); int id = fitness[animal_num-1].id; double x = DNA_result[id].translated_result[0]; double y = DNA_result[id].translated_result[1]; cout<<"最优结果:"<<endl; cout<<"x: "<<x<<" "<<"y: "<<y<<endl; cout<<f(x, y); return 0; }
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
- 87
- 88
- 89
- 90
- 91
- 92
- 93
- 94
- 95
- 96
- 97
- 98
- 99
- 100
- 101
- 102
- 103
- 104
- 105
- 106
- 107
- 108
- 109
- 110
- 111
- 112
- 113
- 114
- 115
- 116
- 117
- 118
- 119
- 120
- 121
- 122
- 123
- 124
- 125
- 126
- 127
- 128
- 129
- 130
- 131
- 132
- 133
- 134
- 135
- 136
- 137
- 138
- 139
- 140
- 141
- 142
- 143
- 144
- 145
- 146
- 147
- 148
- 149
- 150
- 151
- 152
- 153
- 154
- 155
- 156
- 157
- 158
- 159
- 160
- 161
- 162
- 163
- 164
- 165
- 166
- 167
- 168
- 169
- 170
- 171
- 172
- 173
- 174
- 175
- 176
- 177
- 178
- 179
- 180
运行结果:

看得出和python运行出的结果是差不多的。
总结
通过先前python语言的遗传算法实现以及现在c语言遗传算法的实现,我感觉能够比较好的理解遗传算法的思路了,如果认真看完我这两篇博客,一个也能够比较好的理解遗传算法了吧,但是主要还是需要自己敲敲代码,懂得会比较快。
声明:本文内容由网友自发贡献,不代表【wpsshop博客】立场,版权归原作者所有,本站不承担相应法律责任。如您发现有侵权的内容,请联系我们。转载请注明出处:https://www.wpsshop.cn/w/IT小白/article/detail/798272?site
推荐阅读
相关标签





